Polyploidy
inferring the presence of polyploidy and its importance in plants
Diversification, key innovation, and the grand challenges
Polyploidy was long appreciated in plants, but took on a new importance since the advent of genomics as it simultaneously became a source of novelty and a bioinformatic challenge. I have worked on methods for detecting ancient whole-genome duplications (WGDs) with distributions of evolutionary distances between paralogs within a species 1 or distributions of gene count data 2 or gene trees 3 to place WGDs in a phylogenetic context. I am broadly interested in the consequences of polyploidy on genome architecture and gene family evolution.
I am currently investigating how phasing data in polyploids can be useful for population genomics and inferring phylogenetic networks. This includes developing methods and pipelines for phasing 4, and how to apply those phased data to demographic analyses under the coalescent. Polyploids were usually tossed from population genetic studies or dealt with in a way that makes strong assumptions about the polyploidy mechanism. My goal is to get everybody to the haploid level so we can work with mixed ploidy systems under a unified coalescent framework. We are just scratching the surface of developing and applying these methods, but simulations suggest there can be large benefits to phasing on network estimation with allopolyploids.
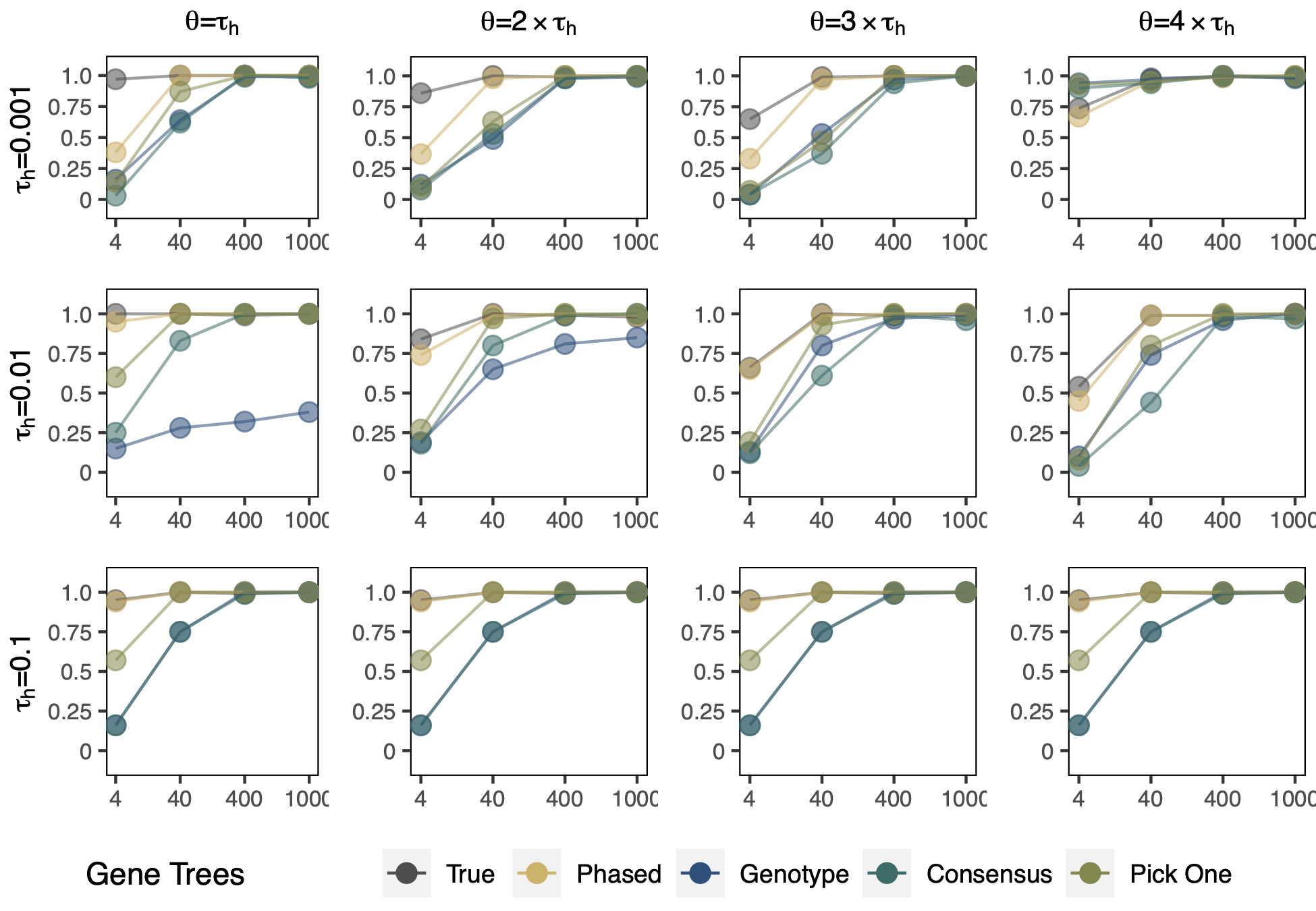 Figure 2 from Tiley et al. 2021 4 Simulation results for whether a network is correct given different treatments of the sequence data as phased as unphased. Either having all of the phased haplotypes (phased) or simply having one phased haplotype sampled per locus (pick one) seems to do well across all simulation conditions with 40 or more loci. A consensus approach also works well once enough loci are sampled. Treating allopolyploid data as an unphased diploid (genotype) can be problematic even as the number of loci increase though.
Figure 2 from Tiley et al. 2021 4 Simulation results for whether a network is correct given different treatments of the sequence data as phased as unphased. Either having all of the phased haplotypes (phased) or simply having one phased haplotype sampled per locus (pick one) seems to do well across all simulation conditions with 40 or more loci. A consensus approach also works well once enough loci are sampled. Treating allopolyploid data as an unphased diploid (genotype) can be problematic even as the number of loci increase though.
The utility of genotyping and phasing polyploids is not yet clear, at least to me. Will correctly genotyping individuals at various ploidy levels lead to better estimates of site frequency spectra and ultimately more accurate model inferences? I am working on these areas now as part of MADGRASS.
-
Tiley GP, Barker MS, Burleigh JG. 2018. Assessing the Performance of Ks Plots for Detecting Ancient Whole Genome Duplications. Genome Biology and Evolution 10:2882-2898. ↩
-
Tiley GP, Ané C, Burleigh JG. 2016. Evaluating and characterizing ancient whole-genome duplications in plants with gene count data. Genome Biology and Evolution 8:1023-1037. ↩
-
Li Z†,Tiley GP†, Galuska SR, Reardon CR, Kidder TI, Rundell RJ, Barker MS. 2018. Multiple large-scale gene and genome duplications during the evolution of hexapods. PNAS 115:4713-4718. ↩
-
Tiley GP, Crowl AA, Manos PS, Sessa EB, Solís-Lemus C, Yoder AD, Burleigh JG. 2021. Phasing alleles improves network inference with allopolyploids bioRxiv doi: https://doi.org/10.1101/2021.05.04.442457
↩ ↩2