Mutation Rate Variation
estimating spontaneous mutation rates from whole-genome sequences
Variation of rates and spectra between and within species
Very recently it has become feasible to characterize the mutations that arise in a single generation by deep sequencing of whole genomes from pedigrees. When parents and their offspring are available, one can simply count the mutations and what type of mutations are in the offspring but absent from the parents. There are a number of caveats though, reviewed in Yoder and Tiley (2021) 1.
Measuring de novo mutation rates can yield quite surprising patterns that invoke the need for deeper understandings of lineage-specific differences in DNA repair mechansism, as revealed by our investigation in mouse lemurs 2. For example, mouse lemurs have a reduced number of C->T transitions at CpG sites.
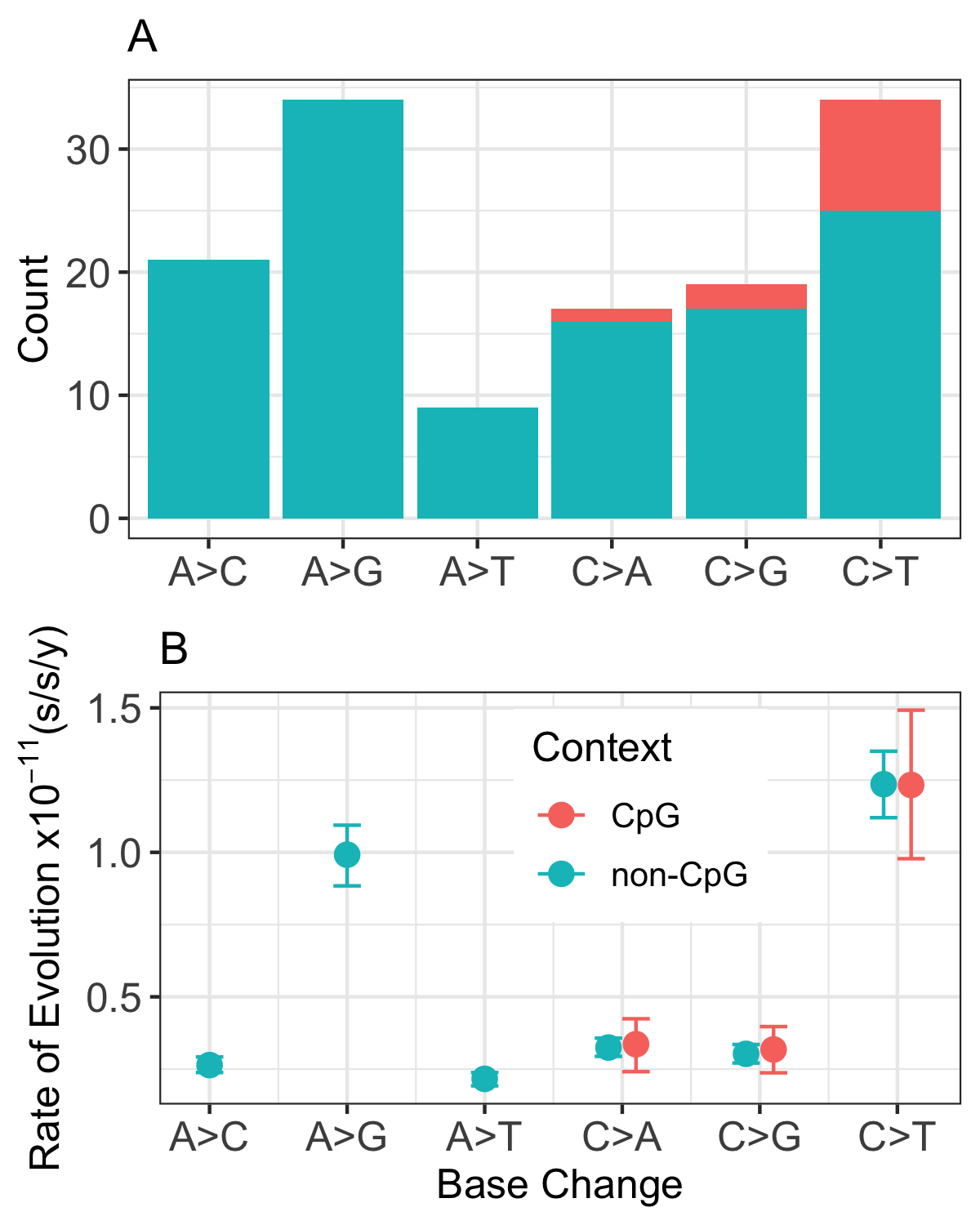
Figure 2 from Campbell, Tiley et al. 2021 2 Most primates have a majority of their C->T transitions in CpG sites, whereas in mouse lemurs, these are the minority (A). While unexpected, the pedigree-based results are supported by phylogenetic relaxed-clock models that partition sites by substitution type. Mouse lemurs show a near-equal rate of C->T transitions at CpG and non-CpG sites (B), while the C->T transition rate at CpG sites is higher in other sampled primates.
I am do not have active research in this area at the moment, but some nice people continue to include me in the conversation 3. I am to pivot from examining species-specific differences to evaluating ecological correlates with mutation rates with population-level data in the near future.
-
Yoder AD, Tiley GP. 2021. The challenge and promise of estimating the de novo mutation rate from whole-genome comparisons among closely related individuals. Molecular Ecology 30:6087-6100. ↩
-
Campbell CR†, Tiley GP†, Poelstra JW, Hunnicutt KE, Larsen PA, Lee H-J, Thorne JL, dos Reis M, Yoder AD. 2021. Pedigree-based and phylogenetic methods support surprising patterns of mutation rate and spectrum in the gray mouse lemur. Heredity 127:233-244. ↩ ↩2
-
Bergeron LA, Besenbacher S, Turner T, Versoza CJ, Wang RJ, Price AL, Armstrong E, Riera M, Carlson J, Chen H-Y, Hahn MW, Harris K, Jleppe AS, López-Nandam EH, Moorjani P, Pfeifer SP, Tiley GP, Yoder AD, Zhang G, Schierup MH. 2022. The mutationathon highlights the importance of reaching standardization in the estimates of pedigree-based germline mutation rates. eLife 11:e73577. ↩