Coalescent Methods
macroevolution as an extension of microevolutionary processes
Divergence times, demographic change, gene flow, and conservation
The multispecies coalescent (MSC) model has been important for species tree estimation where incomplete lineage sorting generates gene tree discordance among species. I am more interested in how the MSC (and coalescent methods more broadly) can be used to infer divergence times between species and populations or change in population size over time.
I am also interested in network estimation under the MSC. The goal is to take a data-inclusive approach, moving from species tree estimation followed by post-hoc tests for introgression to probabilistic inference with networks. I am currently investigating the performance of network methods for estimating speciation times and the timing of introgression or hybridization.
The MSC permeates many aspects of my research including chickens 1, mouse lemurs 2, divergence time estimation 3, and conservation genetics 4. The MSC is incredibly valuable for conservation genetics, as it allows robust estimation of divergence time and population sizes with few (2 or 3) individuals per population compared to allele frequency-based approaches. I still stress that sometimes simpler and less computationally demanding methods work very well though, it depends on the amount of expected ILS
.
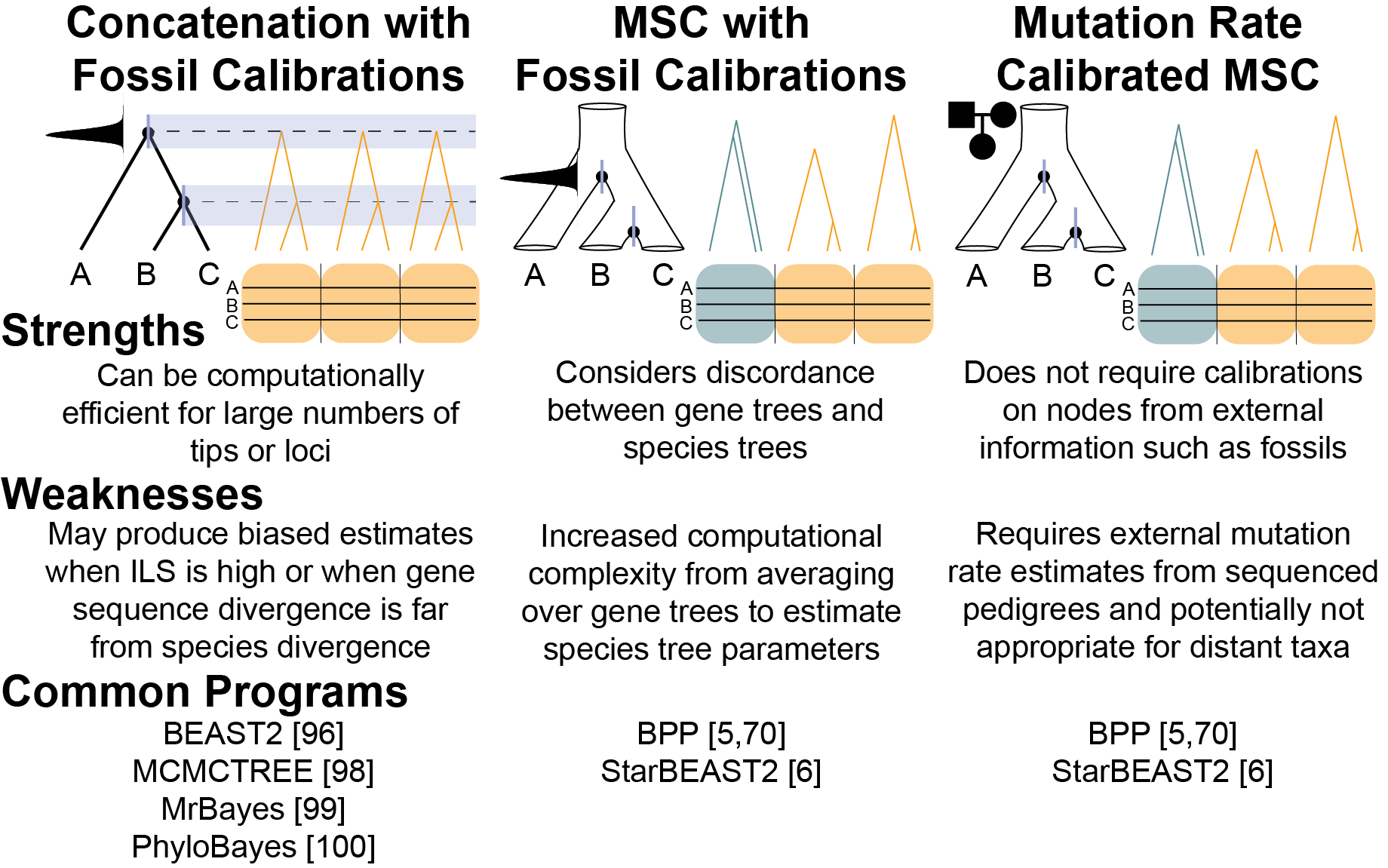 Figure 5 from Tiley et al. 2020b 4 Comparison of different divergence time estimation strategies.
Figure 5 from Tiley et al. 2020b 4 Comparison of different divergence time estimation strategies.
-
Tiley GP, Pandey A, Kimball RT, Braun EL, Burleigh JG. 2020a. Whole genome phylogeny of Gallus: introgression and data-type efects. Avian Research 11:1-15. ↩
-
Poelstra JW†, Salmona J †, Tiley GP†, Schüßler D, Blanco MB, Andriambeloson JB, Bouchez O, Campbell CR, Etter PD, Hohenlohe PA, Hunnicutt KE, Iribar A, Johnson EA, Kappeler PM, Larsen PA, Manzi S, Ralison JM, Randrianambinina B, Rasoloarison RM, Rasolofoson DW, Stahlke AR, Weisrock D, Williams RC, Chikhi L, Louis Jr EE, Radespiel U, Yoder AD. 2020. Cryptic Patterns of Speciation in Cryptic Primates: Microendemic Mouse Lemurs and the Multispecies Coalescent. Systematic Biology 70:203-218.
↩ -
Tiley GP, Poelstra JW, dos Reis M, Yang Z, Yoder AD. 2020b. Molecular Clocks without Rocks: New Solutions for Old Problems. Trends in Genetics 36:845-856.
↩ -
Yoder AD, Poelstra JW, Tiley GP, Williams R. 2018. Neutral Theory is the foundation of Conservation Genetics. Molecular Biology and Evolution 6:1322-1326. ↩ ↩2